X-Ray Imaging
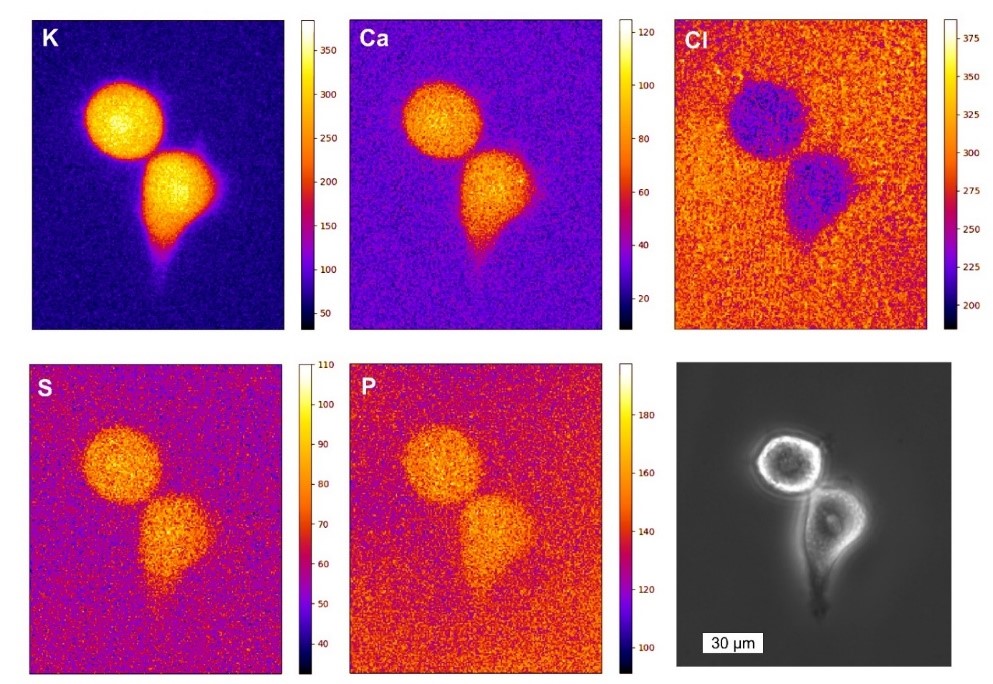
We develop and apply imaging techniques such as X-ray holography,[1] coherent X-ray diffraction imaging,[2] X-ray Ptychography,[3,4] X-ray fluorescence nanoanalysis [5,6], X-ray fluorescence tomography and small angle X-ray scattering [8,9] using radiation from synchrotron sources and free electron lasers. X-ray imaging techniques provide high spatial resolution below 50 nm with intrinsic chemical contrast. In particular for biological samples, micro X-ray fluorescence analysis (µ-XRF) of cryogenically frozen specimen (e.g. transfected cells, diatoms, bacteria, plant material, tissue sections) provides a unique insight into their elemental composition. In collaboration with the P06 team at DESY we develop a new cryogenic X-ray nanoanalysis platform dedicated for the XRF analysis of biological samples. As biological experiments require a good statistical basis to draw solid conclusions, we recently integrated a detector system with high acceptance angle for improved sample throughput.[5]
With this setup, the element distribution in cryogenically prepared HeLa cells was investigated, and physiologically relevant elements such as zinc, chlorine and potassium were imaged.[6] In addition it was investigated how the metal homeostasis was changed if cells overexpress the Htt protein, which causes the formation of amyloid aggregates. This pathway is highly relevant for Huntington’s disease, a fatal neuronal disorder. Increased levels of zinc and potassium were found in HTT expressing cells and the comparison with synthetic intracellular aggregates based on PIC allowed to unravel that the cellular response was a systemic one.[7] The intracellular aggregate-formation was also analyzed by cryo-nano-SAXS and we were able to show the presence of oligomer structures inside the cytosol of the cells. In addition, the structure of repetitive units within the Htt inclusion bodies was retrieved from the SAXS data.[8] The instrument at the P06 beamline is currently extended towards tomographic XRF nanoanalysis to reveal the quantitative cross-sectional element distribution of cryogenic biological material.
X-ray scattering has also been recently introduced as a valuable tool to discover novel antimicrobial modes of action[9]. In a series of experiments, the possibility of distinguishing the mechanisms of action of antibiotics by using scatter patterns of treated E-coli bacteria was proven.[10,11] The method was applied to discover successful candidates within a series of rationally designed hybrid peptides[12] and the technique was extended towards multi-resistant MRSA as clinically relevant strain [13].
Students working on this project: Lejla Jusufagic & Dr. Christoph Rumancev
Collaboration with Dr. Schröder, Dr. Garrevoet, Dr. Falkenberg, Prof. Schroer, DESY
Collaboration with Prof. Schroer, DESY Photon Science
Collaboration with Prof. Ebbinghaus, TU Braunschweig
Collaboration with Prof. Huber, Universität Paderborn
Collaboration with Prof. Hilpert, St, Georges University of London
Collaboration with Prof. Metzler-Nolte, Ruhr-Universität Bochum
[1] A. Rosenhahn, R. Barth, F. Staier, T. Simpson, S. Mittler, S. Eisebitt and M. Grunze,
Journal of the Optical Society of America A – Optics Image Science and Vision 2008, 25, 416-422, DOI: 10.1364/JOSAA.25.000416
[2] A. P. Mancuso, T. Gorniak, F. Staier, O. M. Yefanov, R. Barth, C. Christophis, B. Reime, J. Gulden, A. Singer, M. E. Pettit, T. Nisius, T. Wilhein, C. Gutt, G. Grübel, N. Guerassimova, R. Treusch, J. Feldhaus, S. Eisebitt, E. Weckert, M. Grunze, A. Rosenhahn and I. A. Vartanyants, New Journal of Physics 2010, 12, 035003
[3] K. Giewekemeyer, M. Beckers, T. Gorniak, M. Grunze, T. Salditt and A. Rosenhahn,
Optics Express 2011, 19, 1037-1050, DOI: 10.1364/OE.19.001037
[4] M. Beckers, T. Senkbeil, T. Gorniak, M. Reese, K. Giewekemeyer, S. C. Gleber, T. Salditt and A. Rosenhahn, Physical Review Letters 2011, 107, 208101, DOI: 10.1103/PhysRevLett.107.208101
[5] C. Rumancev, A. Gräfenstein, T. Vöpel, S. Stuhr, A. R. von Gundlach, T. Senkbeil, J. Garrevoet, L. Jolmes, B. König, G. Falkenberg, S. Ebbinghaus, W. H. Schroeder, and A. Rosenhahn Journal of Synchrotron Radiation 2020 27, 60–66, DOI: 10.1107/S1600577519014048
[6] C. Rumancev, T. Vöpel, S. Stuhr, A. von Gundlach, T. Senkbeil, S. Ebbinghaus, J. Garrevoet, G. Falkenberg, B. De Samber, L. Vincze, A. Rosenhahn and W. Schroeder, Biointerphases 2021 16, DOI: 10.1116/6.0000593
[7] A. Gräfenstein, C. Rumancev, R. Pollak, B Hämisch, V. Galbierz, W. H. Schroeder, J. Garrevoet, G. Falkenberg, T. Vöpel, K. Huber, S. Ebbinghaus and A. Rosenhahn, ChemistryOpen 2022, 11, DOI: 10.1002/open.202200024
[8] C. Rumancev, T. Vöpel, S. Stuhr, A. von Gundlach, T. Senkbeil, M. Osterhoff, M. Sprung, V. Garamus, S. Ebbinghaus and A. Rosenhahn, ChemSystemsChem 2021, 3, e2000050, DOI: 10.1002/syst.202000050
[9] C. Rumancev, A. Rosenhahn and K. Hilpert, Frontiers in Pharmacology 2022 13:947005, DOI: 10.3389/fphar.2022.947005
[10] A.R. von Gundlach, V.M. Garamus, T. Gorniak, H.A. Davies, M. Reischl, R. Mikut, K. Hilpert and A. Rosenhahn, Biochimica et Biophysica Acta (BBA) – Biomembranes 2016 1858(5), 918-925, DOI: 10.1016/j.bbamem.2015.12.022
[11] A. von Gundlach, V.M. Garamus, T.M. Willey, J. Ilavsky, K. Hilpert and A. Rosenhahn, Journal of Applied Crystallography 2016 1;49(6):2210-2216, DOI: 10.1107/S1600576716018562
[12] K. Hilpert, J. Gani, C. Rumancev, N. Simpson, P. M. Lopez-Perez, V. M. Garamus, A. von Gundlach, P. Markov, M. Scocchi, R. Mikut and A. Rosenhahn, Frontiers in Pharmacology 2021 DOI: 10.3389/fphar.2021.769739
[13] A. von Gundlach, M. P. Ashby, J. Gani, P. M. Lopez-Perez, A. R. Cookson, S. Huws, C. Rumancev, V. M. Garamus, R. Mikut, A. Rosenhahn and K. Hilpert, Frontiers in Pharmacology 2019 DOI: 10.3389/fphar.2019.01127